Benoukraf Lab
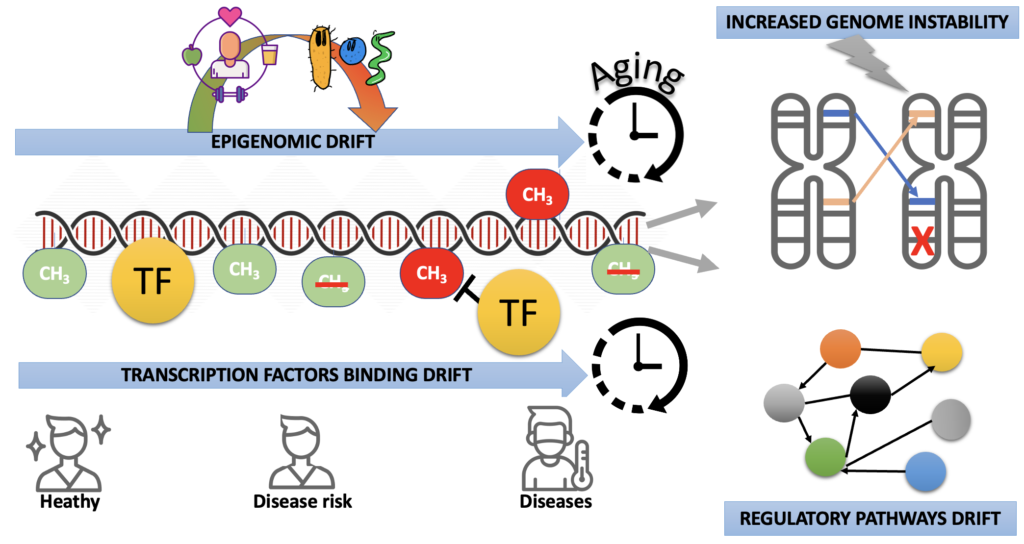
Integrative Omics Approaches to Studying Epigenomic Drift in Disease
Our hybrid laboratory (combining wet and dry methods) focuses on developing computational tools and producing sequencing datasets for analyzing multi-omics interactions in the context of epigenomics drift.
Our research is divided into three main axes:
-
Investigating the chromatin dynamics regulated by transcription factors in the context of epigenomic drift.
-
Investigating the contribution of the microbiome in the epigenomic drift.
-
Investigating the contribution of the epigenomic drift in genome instability.
Overall, using integrative approaches, our laboratory’s goal is to deepen our understanding of epigenomic drift and its underlying mechanisms. By analyzing these different omics layers, we aim to better describe and understand the role of epigenomic drift in pathophysiology.
MethMotif 2024
New release!
A transcription factor binding motifs database coupled with DNA methylation profiles
forkedTF
ForkedTF is an R-library that introduces Forked-PMW and Forked-Sequence Logos to provide a more comprehensive depiction of the sequence affinity of a TF of interest
Human Genetics & Genomics/Cancer & Development Seminar Series
Modules:
MED 6400, 6401, 6402, 6403: Msc students
MED 6410, 6411, 6412, 6413: PhD students
Every Monday at 12:00pm (check D2L for schedule).
Memorial University of Newfoundland
Bioinformatics
Module MED 6393
October 18th 2023,
4.00pm – 5.00pm
October 25th 2023,
2.00pm – 3.45pm
Memorial University of Newfoundland
Impact de la méthylation de l’ADN sur la régulation transcriptionnelle
Biologie computationnelle des systèmes
6 Décembre 2023 Université Saint Joseph de Beyrouth
DNA sequencing technologies
Module CDM5104
January 26th 2024 4.00pm – 6.00pm.
National University of Singapore
Future of Genetics
Class of 2025 / Phase 3 Schedule
Future of Genetics
January 19th, 2024 11.30am (1M101)
Memorial University of Newfoundland
Touati Benoukraf, PhD

Dr. Khadija Rebbani
Senior Post Doctoral Fellow
Transcription factors and DNA methylation interplay

Dr. Akinola A. Alafiatayo
MoH-TFRI / MITACS Post Doctoral Fellow
Omics in Colorectal Cancer

Dr. Hayley Alloway
Post Doctoral Fellow
Chromatin Structure

Zia Hasan
PhD Student (Neuro – Qi lab)
Alzheimer’s single cell omics interplays

Asmaa Mahmoud
PhD student (Genetics)
Characterization of structural variants and repetitive elements
Tian Qin
PhD Student (Neuro – Qi lab)
Dynamics of single-cell brain epigenomics during fear communication

Jason Huan
Professional Associate (Invited)
Full-stack software developer


July 25, 2023 @ 10X Genomics user group meeting | Very proud to see our former students’ success in the industry! Nice job, Quy & Cheng Yong!


July 5-7, 2023 | Congratulations to Asmaa, Matthew, and Gaston for receiving awards during the BioMedicine 2023 symposium!


May 26, 2023 | Very delighted to have shared science with high school students during Discovery Day @ Memorial University. We hope we were able to instill a passion for genomics in these future bright scientists.

April 2023 | Very delighted to meet Fred Fox, Terry Fox‘s brother, who was a Canadian athlete, humanitarian, and cancer research activist. The Terry Fox Research Institute, which was named in honor of Terry Fox, is a key player in cancer research in Canada, and one of our sponsors.

Seminar on structural variations in Leukemia. Invited speakers: Touati Benoukraf.
TAGC, Aix-Marseille University, France | December 16, 2022